Create a heatmap or frequency table from a list of Rxs trees
freqTab_heatMaps.RdThese functions create a heatmap showing the frequencies with which
specific possible entity values occur for the intersection of two entities -
or they create the underlying frequency table for the set of sources or just
one source. You usually only use heatMap_from_rxsProject, which takes the
object with Rxs trees and creates a heatmap.
heatMap_from_freqTab(
freqTab,
rowOrder = rownames(freqTab),
colOrder = colnames(freqTab),
rowLab = "Row",
colLab = "Column",
freqLab = "Frequency",
plotTitle = paste0("Frequency Table for ", rowLab, " and ", colLab),
xLabelRotationAngle = 45,
legend.position = "right",
fillScale = ggplot2::scale_fill_viridis_c(),
theme = ggplot2::theme_minimal()
)
heatMap_from_rxsProject(
x,
rowRegex,
colRegex,
freqTabArgs = NULL,
rowOrder = NULL,
colOrder = NULL,
rowLab = "Row",
colLab = "Column",
freqLab = "Frequency",
plotTitle = paste0("Frequency Table for ", rowLab, " and ", colLab),
xLabelRotationAngle = 45,
legend.position = "right",
flattenValues = TRUE,
fillScale = ggplot2::scale_fill_viridis_c(),
theme = ggplot2::theme_minimal()
)
rxsProject_to_freqTab(
x,
rowRegex,
colRegex,
rowTargetValue = NULL,
colTargetValue = NULL,
fillValue = 0,
rowColMultiplicationFunction = `*`,
rowTargetFunction = `==`,
colTargetFunction = `==`,
aggregationFunction = `+`,
rowLabels = NULL,
colLabels = NULL,
rowOrder = NULL,
colOrder = NULL,
sortRowsAlphabetically = TRUE,
sortColsAlphabetically = TRUE,
includeValueListsOfMatch = TRUE,
flattenValues = TRUE,
excludeParentWhenValueListReturned = TRUE,
silent = metabefor::opts$get("silent")
)
rxsTreeList_to_freqTab(
x,
rowRegex,
colRegex,
rowTargetValue = NULL,
colTargetValue = NULL,
fillValue = 0,
rowColMultiplicationFunction = `*`,
rowTargetFunction = `==`,
colTargetFunction = `==`,
aggregationFunction = `+`,
rowLabels = NULL,
colLabels = NULL,
rowOrder = NULL,
colOrder = NULL,
sortRowsAlphabetically = TRUE,
sortColsAlphabetically = TRUE,
includeValueListsOfMatch = TRUE,
flattenValues = TRUE,
excludeParentWhenValueListReturned = TRUE,
silent = metabefor::opts$get("silent")
)
rxsTree_to_freqTab(
x,
rowRegex,
colRegex,
rowTargetValue = NULL,
colTargetValue = NULL,
valuePreprocessingFunction = vectorValue_to_valueList,
rowColMultiplicationFunction = `*`,
rowTargetFunction = `==`,
colTargetFunction = `==`,
rowLabels = NULL,
colLabels = NULL,
rowOrder = NULL,
colOrder = NULL,
sortRowsAlphabetically = TRUE,
sortColsAlphabetically = TRUE,
includeValueListsOfMatch = TRUE,
flattenValues = TRUE,
excludeParentWhenValueListReturned = TRUE,
silent = metabefor::opts$get("silent")
)Arguments
- freqTab
The frequency table (as produced by
rxsTreeList_to_freqTab).- rowOrder, colOrder
The order of the row and column entities/values.
- rowLab, colLab, freqLab, plotTitle
The labels to use for the rows (y axis), columns (x axis), frequencies (fill), and the plot title to use.
- xLabelRotationAngle
The angel to rotate the labels on the X axis.
- legend.position
The position of the legend.
- fillScale
The fill scale to use.
- theme
The ggplot2 theme to use.
- x
The Rxs object or the Rxs tree for which to produce the frequency table.
- rowRegex, colRegex
Regular expressions used to find the entities that will form the rows or columns
- freqTabArgs
Arguments to pass to the frequency table functions.
- flattenValues
Whether to flatten values that are vectors into single string values that represents those values, or not (in which case they remain vectors, and so single sources occur multiple times in the frequency table).
- rowTargetValue, colTargetValue
Value to consider a 'hit' for the row and column entities
- fillValue
The value to insert in rows or columns that have to be added because of inconsistencies between extraction scripts
- rowColMultiplicationFunction
The function to use to multiply the rows and columns into the matrix
- rowTargetFunction, colTargetFunction
Functions to use to compare, the entity values found for the rows and columns to the target values (e.g.
==,>,<, etc).- aggregationFunction
The function to use to aggregate matrices
- rowLabels, colLabels
A names vector used to replace row and column labels; the indices (element names) should be the entity identifiers and the values the labels to use.
- sortRowsAlphabetically, sortColsAlphabetically
Whether to sort columns or rows alphabetically.
- includeValueListsOfMatch
Whether to also include the value lists inside matching entities (useful for quickly selecting e.g. all results)
- excludeParentWhenValueListReturned
Whether, if an entity matches, has a value list as value, and those value lists are returns (i.e.
includeValueListsOfMatchisTRUE), the parent entity (that matched the regular expression) should be excluded.- silent
Whether to be silent or chatty.
- valuePreprocessingFunction
The function to use to preprocess values - only for advanced use.
Value
A ggplot or a frequency table
Details
The underlying frequency table is produced in a number of steps. First,
the values of the entities matching the regular expressions are obtained.
Second, the row and column target functions are applied to these values (as
first argument) and to the specified row and column target values (as second
argument). Then, the rowColMultiplicationFunction is applied to the
result to obtain the value for the cell. Finally, if rxs_to_freqTab
is called, aggregationFunction is called on the frequency tables of the
separate sources.
Examples
### Load an example Rxs Project
example_rxsProject_1 <- metabefor::example_rxsProject_1;
### Create a heatmap
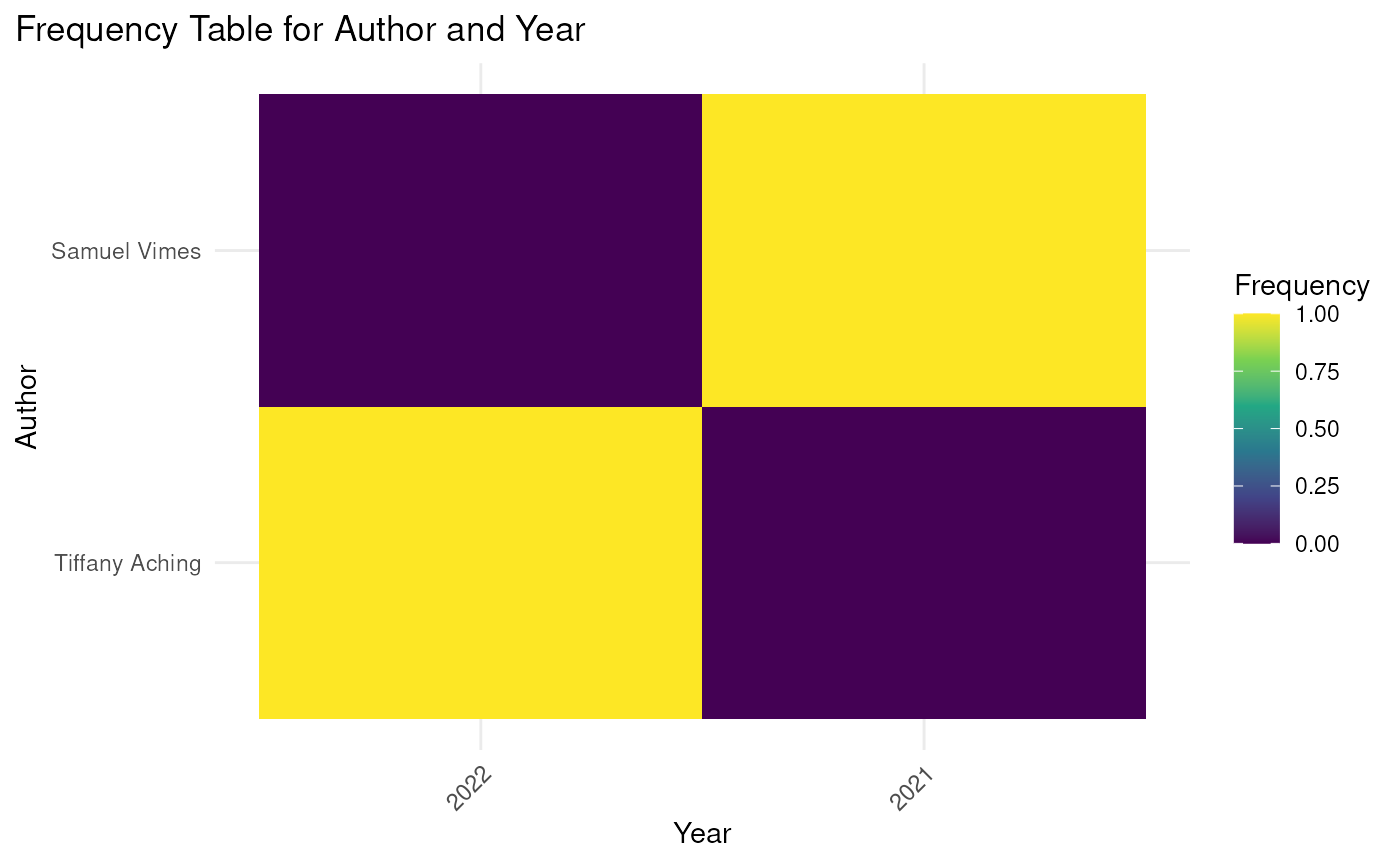
metabefor::heatMap_from_rxsProject(
example_rxsProject_1,
"sourceAuthors",
"publicationYear",
rowLab = "Author",
colLab = "Year"
);
 ### Load an example Rxs Project
example_rxsProject_1 <- metabefor::example_rxsProject_1;
### Create a crosstable with frequencies
metabefor::rxsProject_to_freqTab(
example_rxsProject_1,
"sourceAuthors",
"publicationYear"
);
#> 2022 2021
#> Tiffany Aching 1 0
#> Samuel Vimes 0 1
### Load an example Rxs Project
example_rxsProject_1 <- metabefor::example_rxsProject_1;
### Create a crosstable with frequencies
metabefor::rxsProject_to_freqTab(
example_rxsProject_1,
"sourceAuthors",
"publicationYear"
);
#> 2022 2021
#> Tiffany Aching 1 0
#> Samuel Vimes 0 1